
Ergodicity and model quality in template-restrained canonical and temperature/Hamiltonian replica exchange coarse-grained molecular dynamics simulations of proteins,Journal of Computational Chemistry - X-MOL
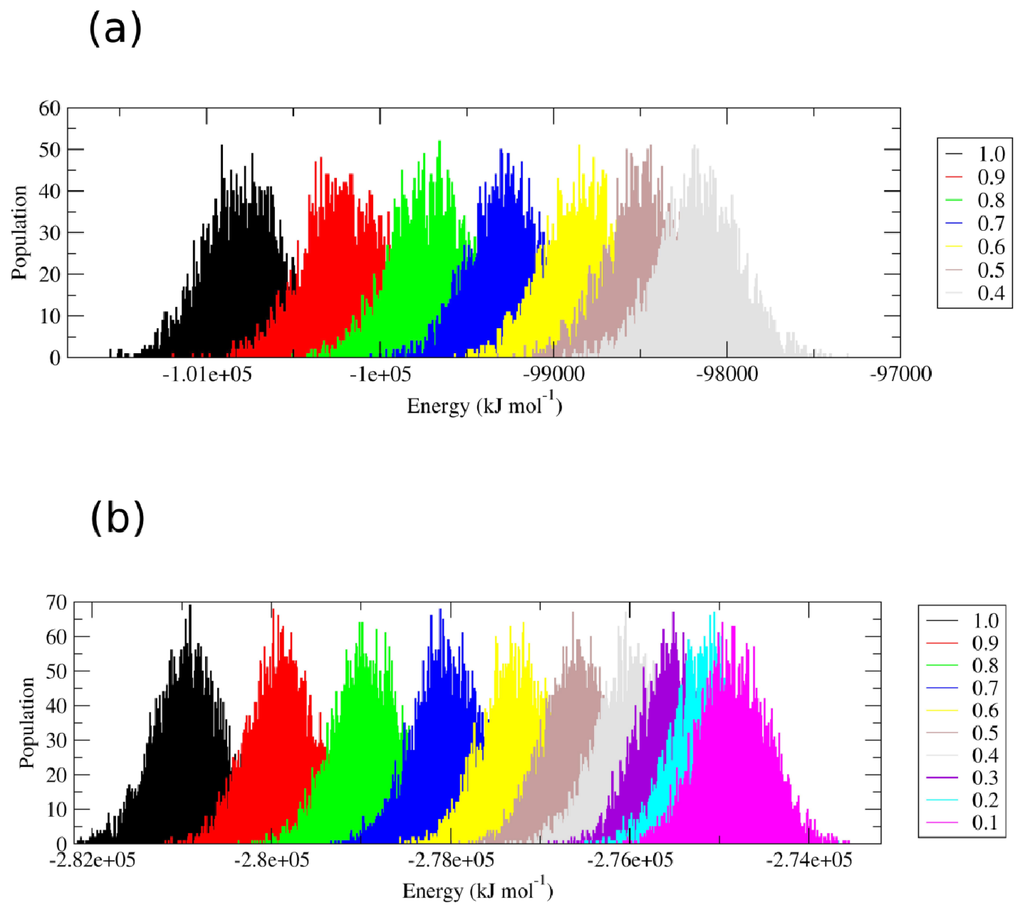
IJMS | Free Full-Text | A Hamiltonian Replica Exchange Molecular Dynamics (MD) Method for the Study of Folding, Based on the Analysis of the Stabilization Determinants of Proteins

Efficient and accurate calculation of proline cis/trans isomerization free energies from Hamiltonian replica exchange molecular dynamics simulations: Structure
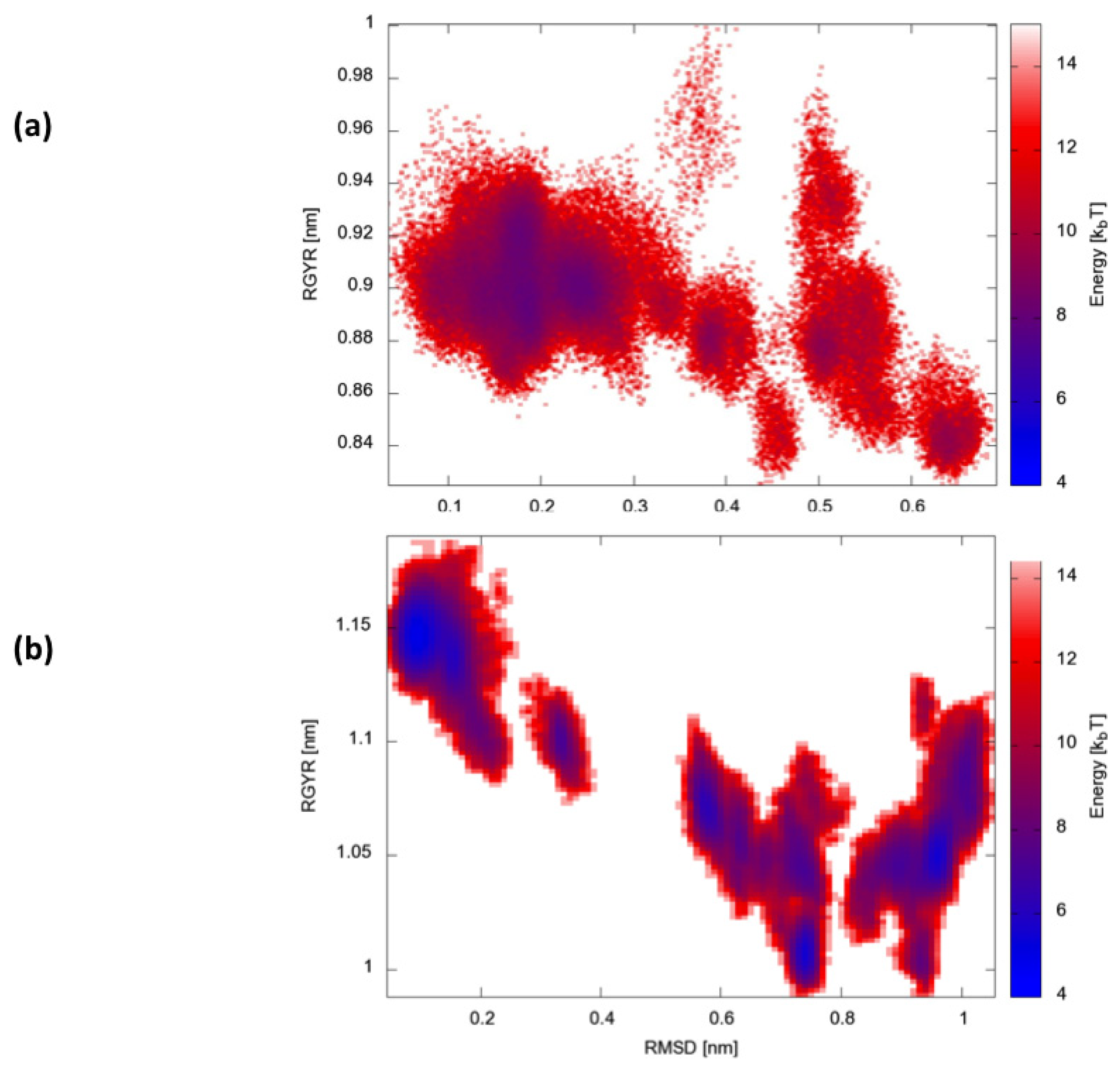
IJMS | Free Full-Text | A Hamiltonian Replica Exchange Molecular Dynamics (MD) Method for the Study of Folding, Based on the Analysis of the Stabilization Determinants of Proteins

Advanced replica-exchange sampling to study the flexibility and plasticity of peptides and proteins. | Semantic Scholar
Recovering Protein Thermal Stability Using All-Atom Hamiltonian Replica- Exchange Simulations in Explicit Solvent

Replica Exchange Molecular Dynamics: A Practical Application Protocol with Solutions to Common Problems and a Peptide Aggregation and Self-Assembly Example. - Abstract - Europe PMC

Molecules | Free Full-Text | Does Hamiltonian Replica Exchange via Lambda-Hopping Enhance the Sampling in Alchemical Free Energy Calculations?
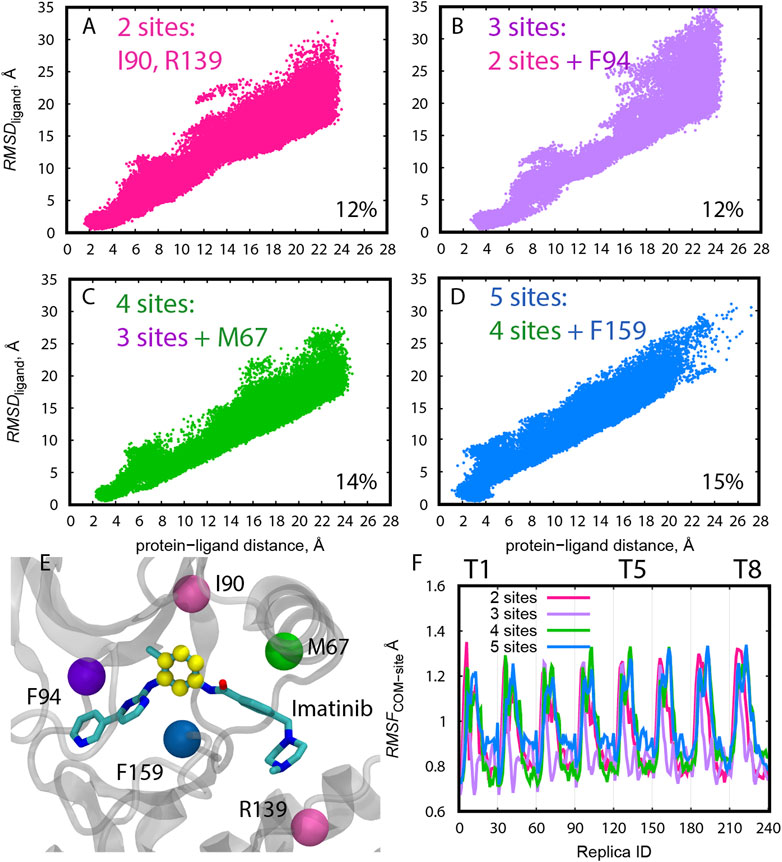
Frontiers | Practical Protocols for Efficient Sampling of Kinase-Inhibitor Binding Pathways Using Two-Dimensional Replica-Exchange Molecular Dynamics
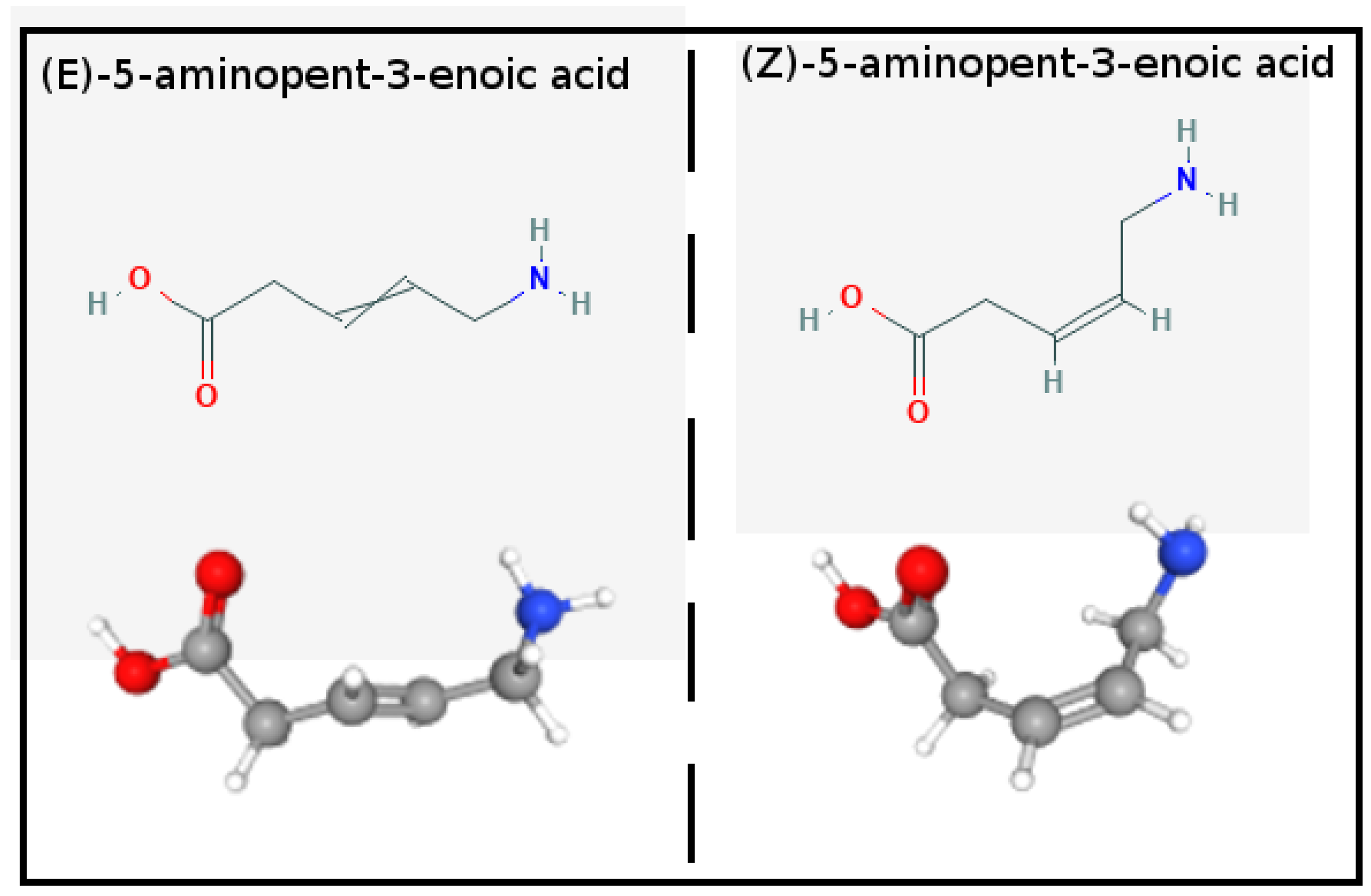
Molecules | Free Full-Text | Does Hamiltonian Replica Exchange via Lambda-Hopping Enhance the Sampling in Alchemical Free Energy Calculations?
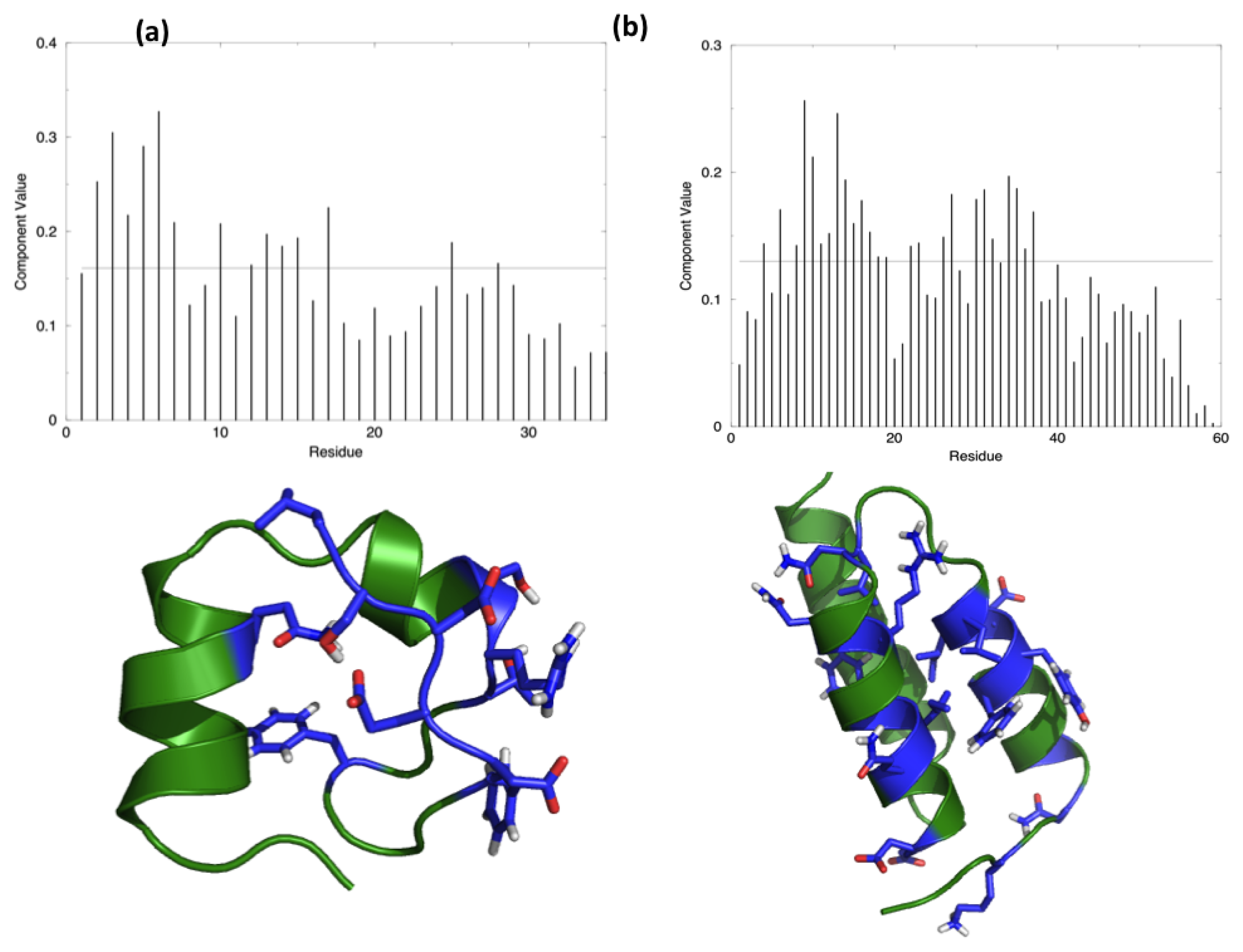
IJMS | Free Full-Text | A Hamiltonian Replica Exchange Molecular Dynamics (MD) Method for the Study of Folding, Based on the Analysis of the Stabilization Determinants of Proteins
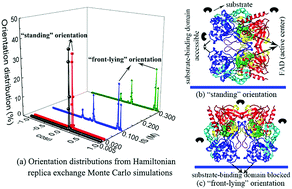
Hamiltonian replica exchange simulations of glucose oxidase adsorption on charged surfaces - Physical Chemistry Chemical Physics (RSC Publishing)

Schematic diagram of different parallel tempering simulations. (a) In... | Download Scientific Diagram
Enhanced Sampling approaches in protein docking. 1. Temperature replica... | Download Scientific Diagram

Computing Alchemical Free Energy Differences with Hamiltonian Replica Exchange Molecular Dynamics (H-REMD) Simulations. - Abstract - Europe PMC
GitHub - huhlim/H-REMD: Hamiltonian and Temperature Replica Exchange Molecular Dynamics simulation using OpenMM
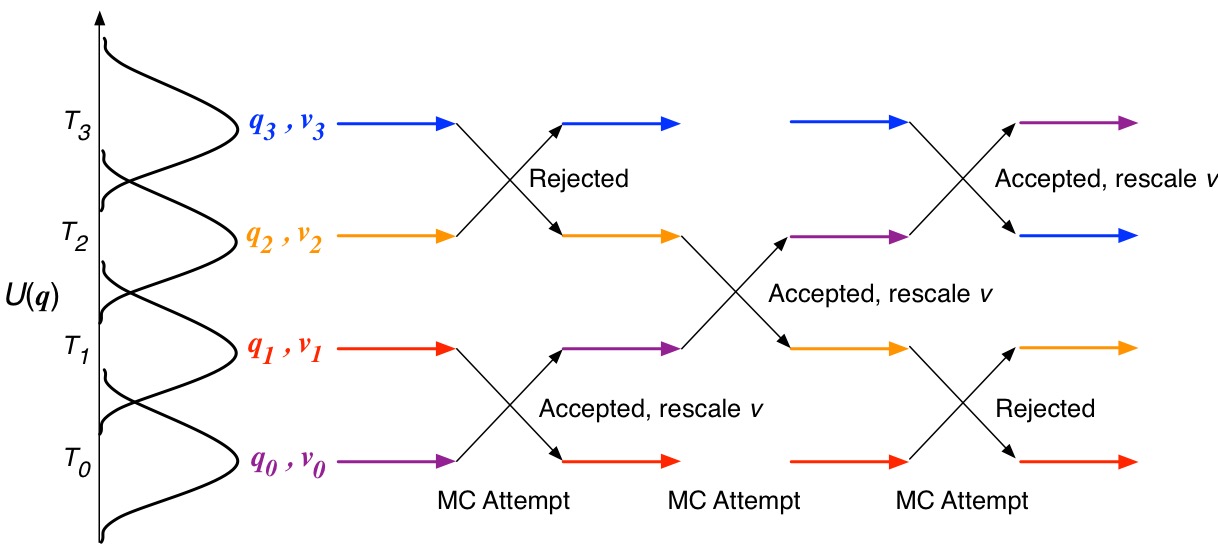

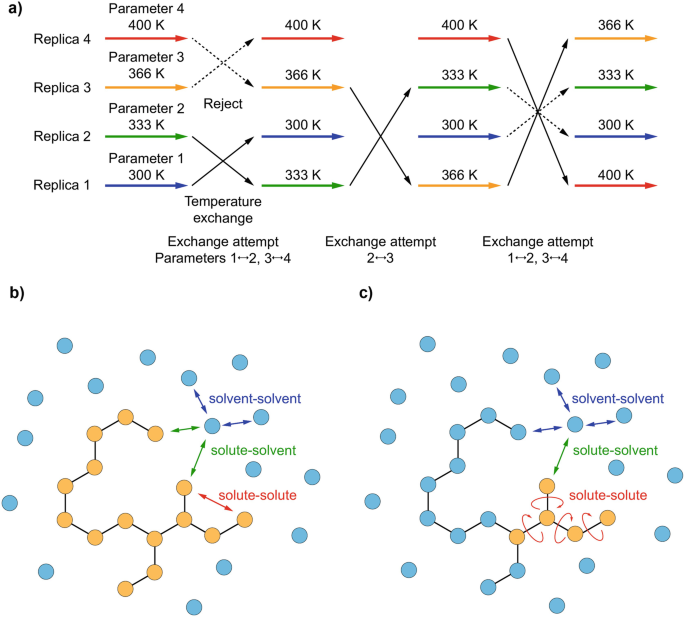



![ASAP] Hamiltonian-Reservoir Replica Exchange and Machine Learning P ASAP] Hamiltonian-Reservoir Replica Exchange and Machine Learning P](https://www.researcher-app.com/image/eyJ1cmkiOiJodHRwczovL3MzLWV1LXdlc3QtMS5hbWF6b25hd3MuY29tL3N0YWNrYWRlbWljL3Byb2R1Y3Rpb24vcGFwZXIvMzU0MTczMi5wbmciLCJmb3JtYXQiOiJ3ZWJwIiwicXVhbGl0eSI6MTAwLCJub0NhY2hlIjp0cnVlfQ==.webp)

