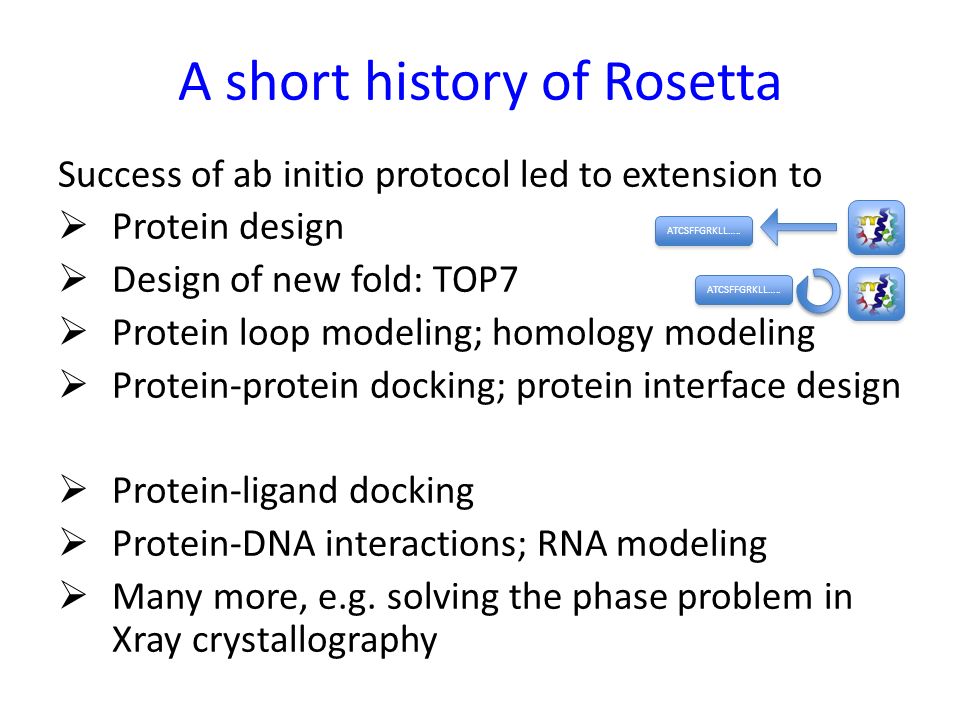
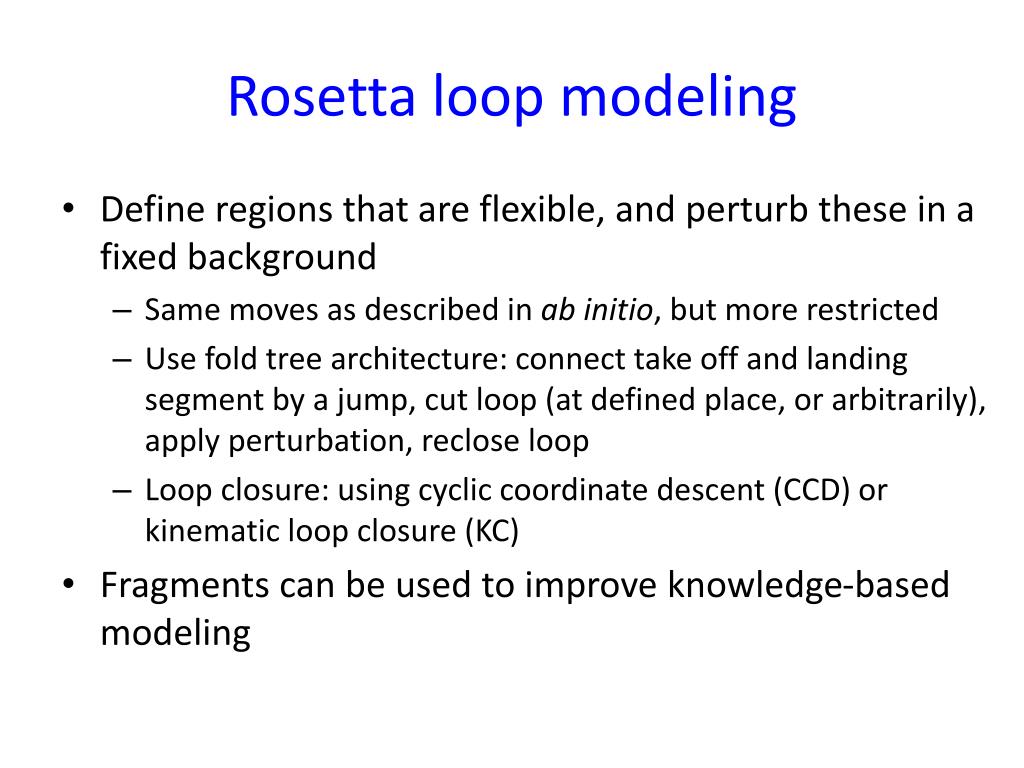
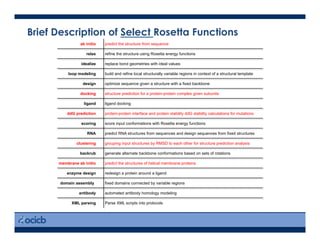
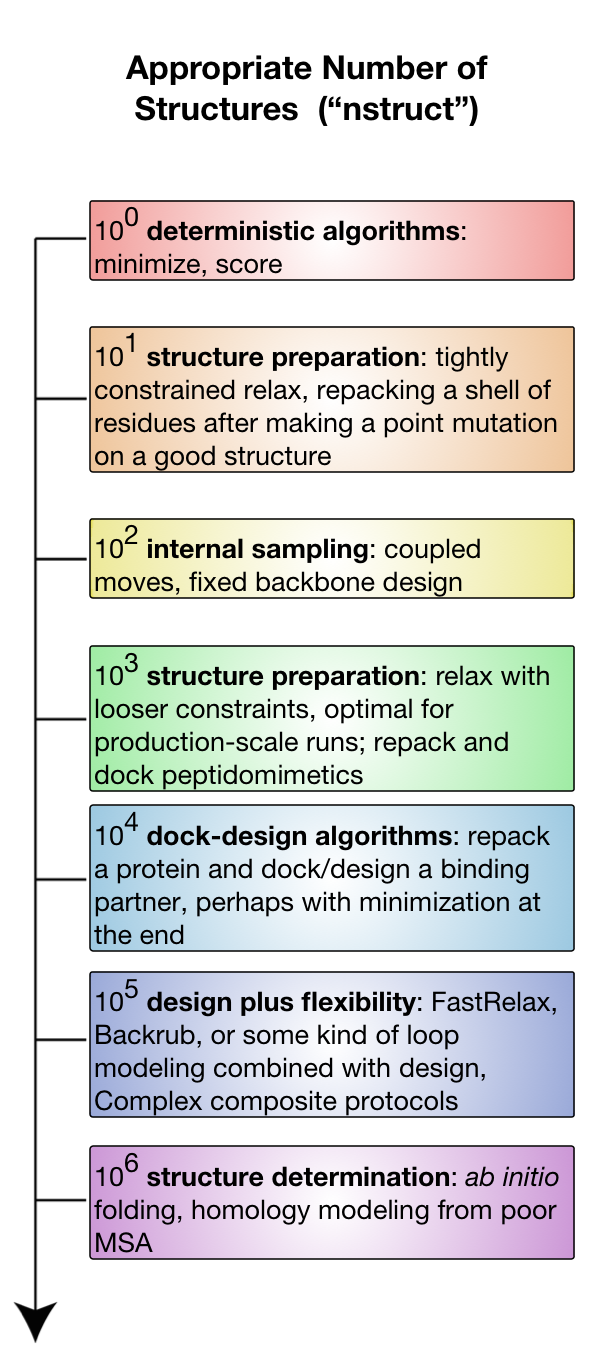
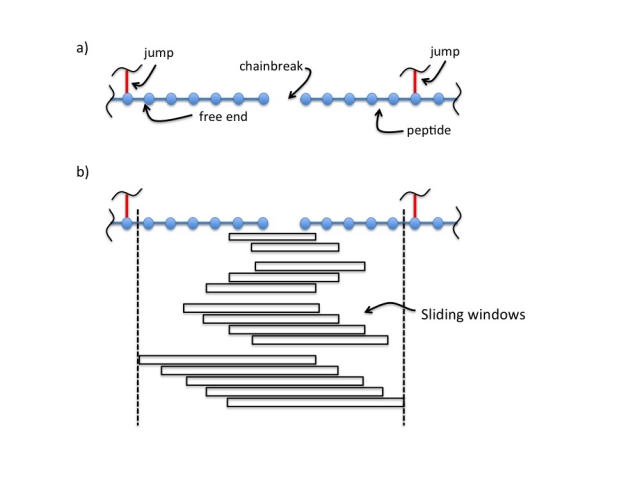
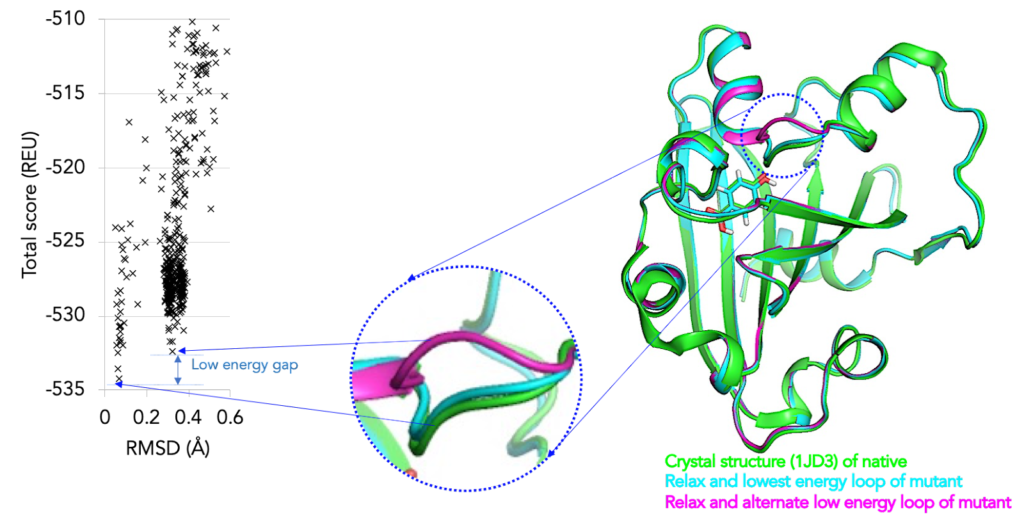
Systematic comparison of Amber and Rosetta energy functions for protein structure evaluation. | Theoretical and Computational Chemistry | ChemRxiv | Cambridge Open Engage
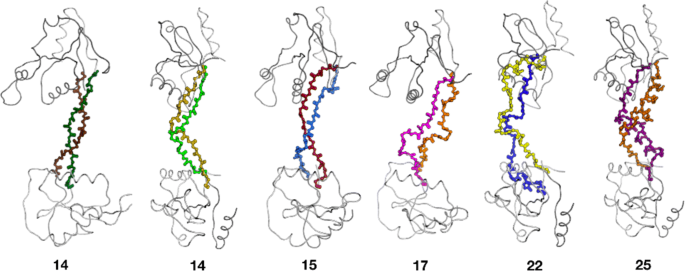
Fast design of arbitrary length loops in proteins using InteractiveRosetta | BMC Bioinformatics | Full Text
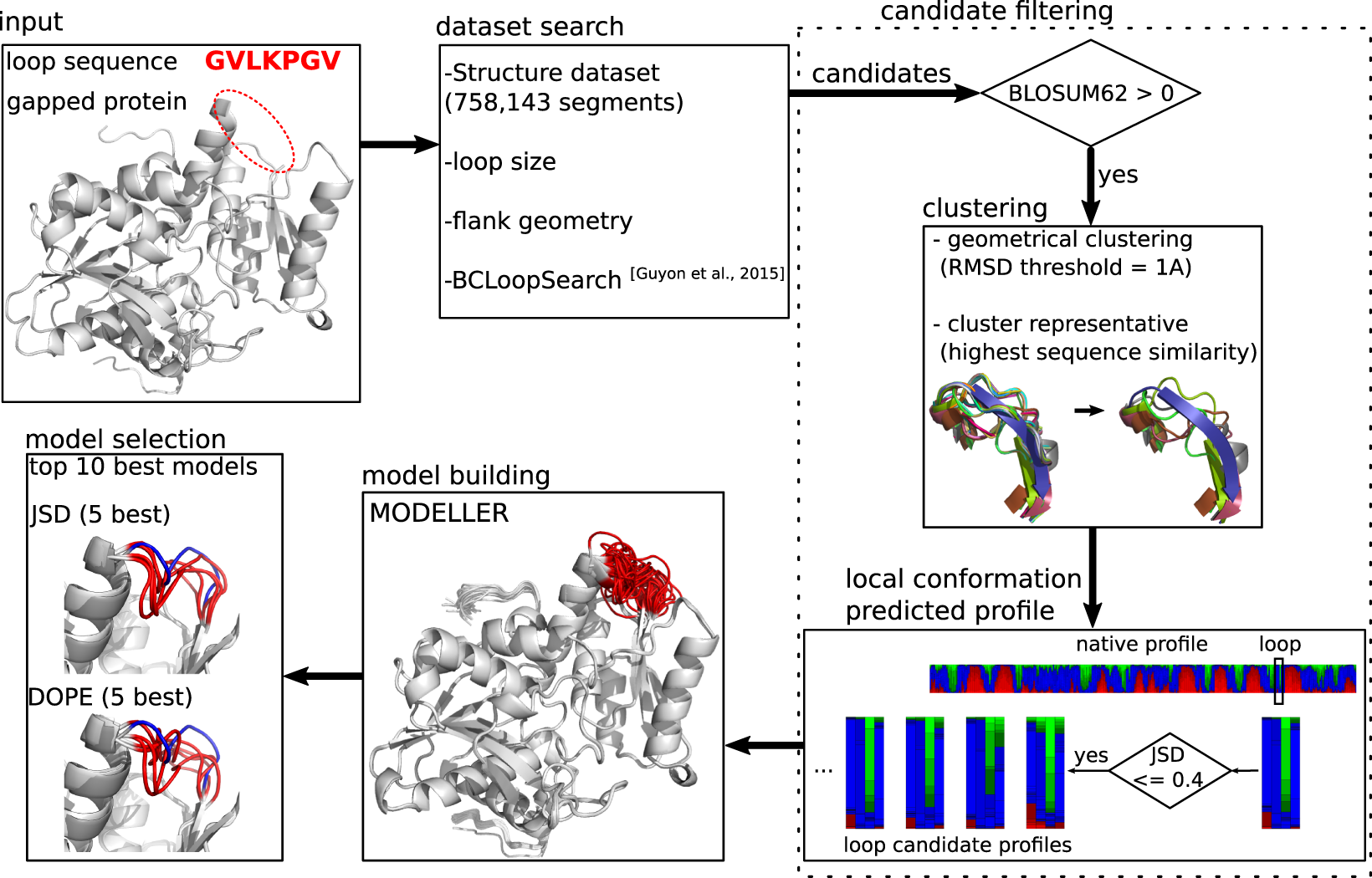
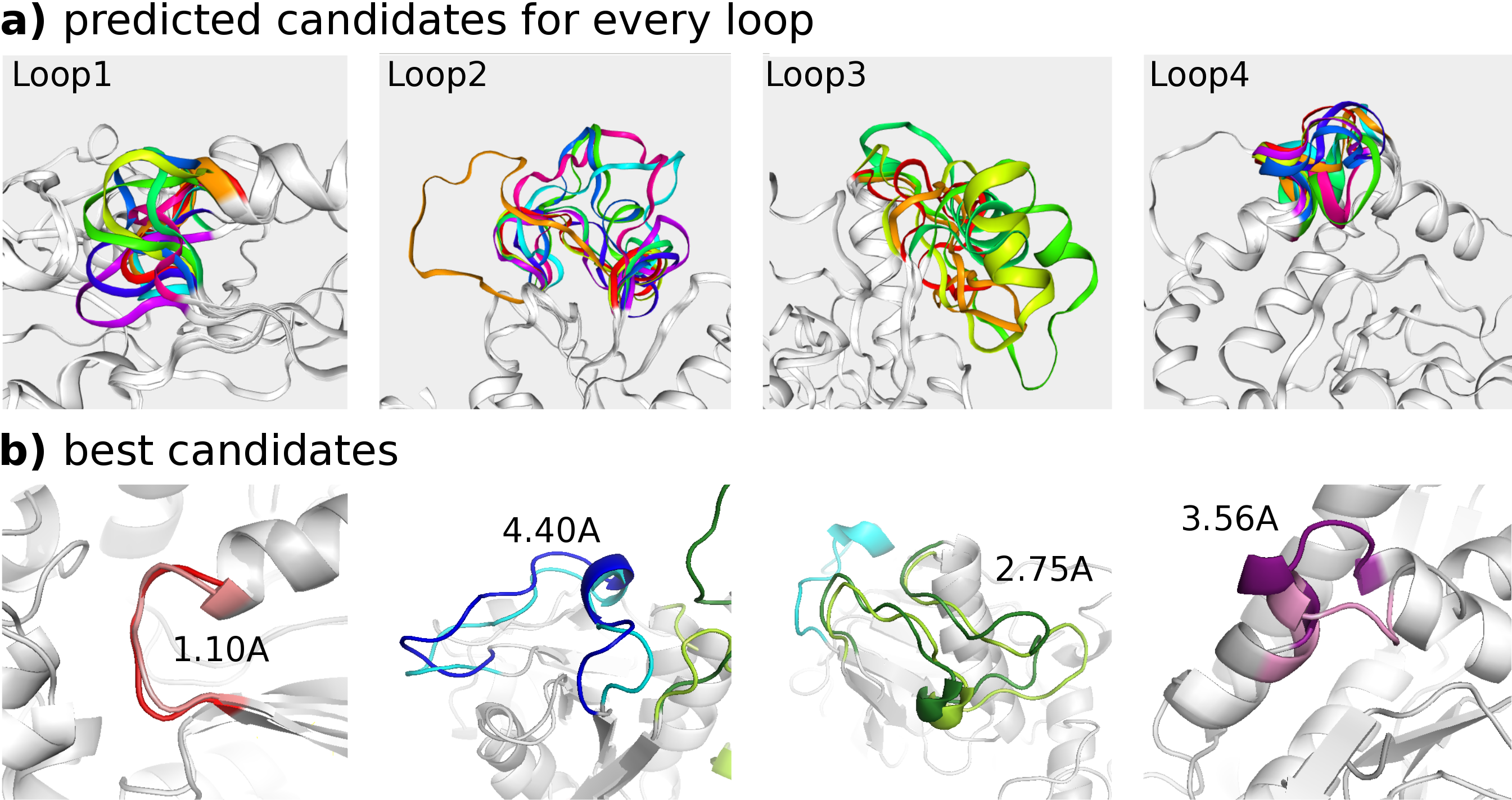
DaReUS-Loop: accurate loop modeling using fragments from remote or unrelated proteins | Scientific Reports

Modeling of protein conformational changes with Rosetta guided by limited experimental data - ScienceDirect

Comparison of experimental and CS-Rosetta-RNA models for diverse RNA... | Download Scientific Diagram
![PDF] PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta | Semantic Scholar PDF] PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/b1d623c7aafa02487d038af433a60c0a444a9db6/2-Figure1-1.png)
PDF] PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta | Semantic Scholar